Hidden Markov Model
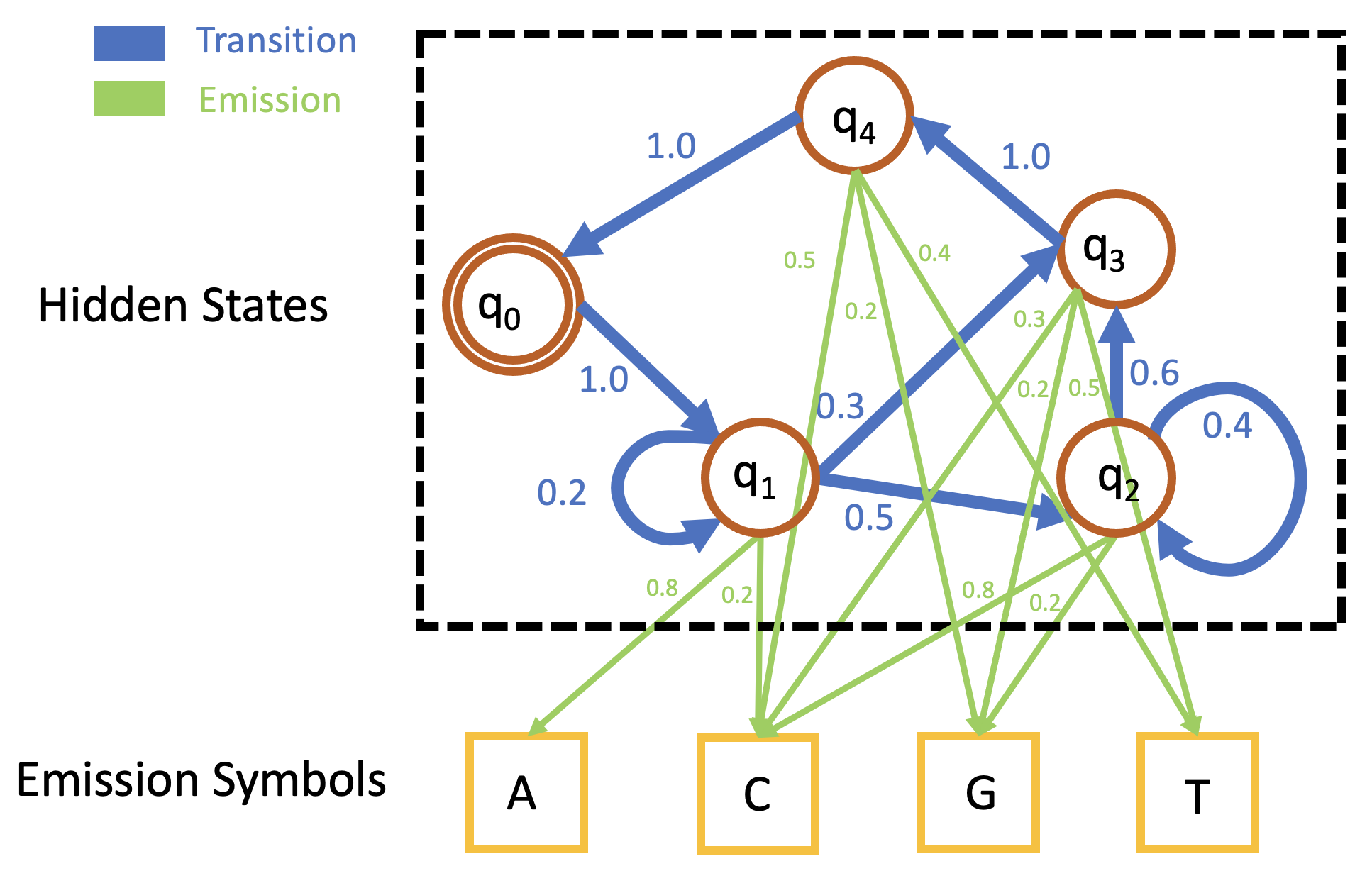
Viterbi Algorithm
Viterbi Algorithm applies dynamic programming to find the optimal path of states to maximize the probability of generating the given sequence. In detail, assume the set of all states is \[\mathcal{S} = \{q_0,q_1,\cdots,q_n\},\] the given sequence is \[L = x_0x_1\cdots x_{l-1},\] our goal is to find the path of states $P$ s.t. \[P = \underset{Q\in \text{all paths}}{\operatorname{arg max}} \,\mathbb{P}(L|Q) = \underset{Q\in \text{all paths}}{\operatorname{arg max}} \,\mathbb{P}_t(q_{0}\,|\,q_{k_{l-1}})\mathbb{P}_t(q_{k_0}\,|\,q_0)\mathbb{P}_e(x_{0}\,|\,q_{k_{0}}) \prod_{i=1}^{l-1} \mathbb{P}_t(q_{k_{i}}\,|\,q_{k_{i - 1}})\mathbb{P}_e(x_{i}\,|\,q_{k_{i}}),\] where $Q= q_{0}q_{k_0}q_{k_1}\cdots q_{k_{l-1}}q_0$. Denote the maximal probability as \[\mathbb{P}_m = \max_{Q\in \text{all paths}} \,\mathbb{P}(L|Q).\] Viterbi algorithm uses the following recursive equation to calculate $\mathbb{P}_m$ and find the path $P$ by tracing back, \[F(i,j)=\begin{cases} \displaystyle\max_{q_s\in \mathcal{S}}F(s,j-1)\mathbb{P}_t(i\,|\,q_s)\mathbb{P}_e(x_j\,|\,q_i)&1\leq j\leq l-1\\ \mathbb{P}_t(q_i\,|\,q_0)\mathbb{P}_e(x_0\,|\,q_i)&j=0\end{cases}\] where $F(i,j)$ is the function of the i-th state ($q_{i}$) and the j-th position in the sequence ($x_{j}$). Since the final step should be the transition to $q_0$, the maximal probability is \[\mathbb{P}_m(L) = \max_{q_s\in \mathcal{S}}F(s,l-1)\mathbb{P}_t(q_0\,|\,q_s).\]
Implementation
Below is the implementation of Viterbi Algorithm.
import numpy as np
def DecodeHMM(HMM, Input_seq):
State = list(HMM.keys())
m = len(Input_seq) #length of seq
n = len(State) #number of states
#Initialization
Score = [[-np.inf for x in range(n)] for x in range(m)] #scoring matrix Score[seq][state]
Pointer =[['Null' for x in range(n)] for x in range(m)] #pointer matrix
for i in range(1,n):
PrTr = HMM[State[0]]['TR'][State[i]]
PrEm = HMM[State[i]]['EM'][Input_seq[0]]
Pr = np.log(PrTr)+np.log(PrEm)
Score[0][i] = Pr
if Pr > -np.inf : Pointer[0][i] = State[0]
#Fill the score matrix and pointer matrix
for pos in range(1,m):
Nuc = Input_seq[pos]#nucleotide
StateEmPos = [ i for i in State if HMM[i]['EM'][Nuc] > 0] #set of states that can emit current nucleotide
for q in StateEmPos:
StateTrq = [i for i in State if HMM[i]['TR'][q] > 0] #set of states that can transit to current state
PrEm = HMM[q]['EM'][Nuc]
for p in StateTrq:
PrTr = HMM[p]['TR'][q]
value = Score[pos-1][State.index(p)] + np.log(PrTr)+np.log(PrEm)
if value > Score[pos][State.index(q)]:
Score[pos][State.index(q)] = value
Pointer[pos][State.index(q)] = p
#trace back
Best = State[1]
Output = [State[0]]
for i in range(2,n):
CurrentScore = Score[m-1][i] + np.log(HMM[State[i]]['TR'][State[0]])
MaxScore = Score[m-1][State.index(Best)] + np.log(HMM[Best]['TR'][State[0]])
if CurrentScore > MaxScore :
Best = State[i]
MaxScore = Score[m-1][State.index(Best)] + np.log(HMM[Best]['TR'][State[0]])
Output.append(Best)
pos = m - 1
while pos != -1:
Best = Pointer[pos][State.index(Best)]
Output.append(Best)
pos -= 1
if MaxScore == - np.inf:
Output = 'Null'
else : Output = Output[::-1]
return MaxScore,Output
For example, we can input a HMM and run the decoding function:
HMM = {'q0':{'TR':{'q0':0,'q1':0.7,'q2':0.2,'q3':0, 'q4':0.1},'EM':{'A':0,'T':0,'C':0,'G':0}},
'q1':{'TR':{'q0':0,'q1':0.2,'q2':0.3,'q3':0.5,'q4':0}, 'EM':{'A':0.45,'T':0.45,'C':0.05,'G':0.0}},
'q2':{'TR':{'q0':0,'q1':0, 'q2':0.8,'q3':0.2,'q4':0}, 'EM':{'A':0.05,'T':0.05,'C':0.25,'G':0.65}},
'q3':{'TR':{'q0':0,'q1':0, 'q2':0.5,'q3':0,'q4':0.5}, 'EM':{'A':0.2,'T':0.6,'C':0.1,'G':0.1}},
'q4':{'TR':{'q0':0.3,'q1':0.2,'q2':0.5,'q3':0,'q4':0},'EM':{'A':0.4,'T':0.1,'C':0.1,'G':0.4}}}
DecodeHMM(HMM, 'ACCCT')
The output is:
(-13.466615801344505, ['q0', 'q1', 'q2', 'q2', 'q3', 'q4', 'q0'])